LoRDEC: hybrid correction of long reads 
Table of Contents
Date : 09-09-2020
News
LoRDEC has been accepted by the Elixir Service Delivery Plan in 2019. LoRDEC is now registered in the ELIXIR bio.tools database (https://bio.tools/ under this entry), and documented following the EDAM ontology. Elixir is a European organisation that coordinates life science resources from across Europe, and the Institut Français de Bioinformatique is the French node of Elixir.

Overview
LoRDEC is a bioinformatics software developed in 2014 at the CNRS, Université de Montpellier (France) in collaboration with Leena Salmela of University of Helsinki (Finland).
LoRDEC processes data coming from high throughput sequencing machines of the second and third generations. These data are called sequencing reads, or simply reads for short. Technically speaking it processes short reads and long reads to correct errors in the long reads.
Third generation DNA sequencing technologies yield long, but error rich sequencing reads, while second generation technologies yield short read with low error rate. Hence, the need for correcting the long reads.
LoRDEC is a program for error correcting long sequencing reads using short reads. It implements a hybrid correction approach. It uses little memory and is very efficient. Most importantly it scales up to process very large data sets. It can be applied to long reads obtained with either Pacific Biosciences SMRT sequencing (SMRT = Single Molecule Real Time) or with Oxford Nanopore MINion technology.
This page maintains up to date information about LoRDEC software, including a table of completed genomic and transcriptomic projects (Table 1), with their distribution grouped by species (Figure 2) and by country (Figure 3).
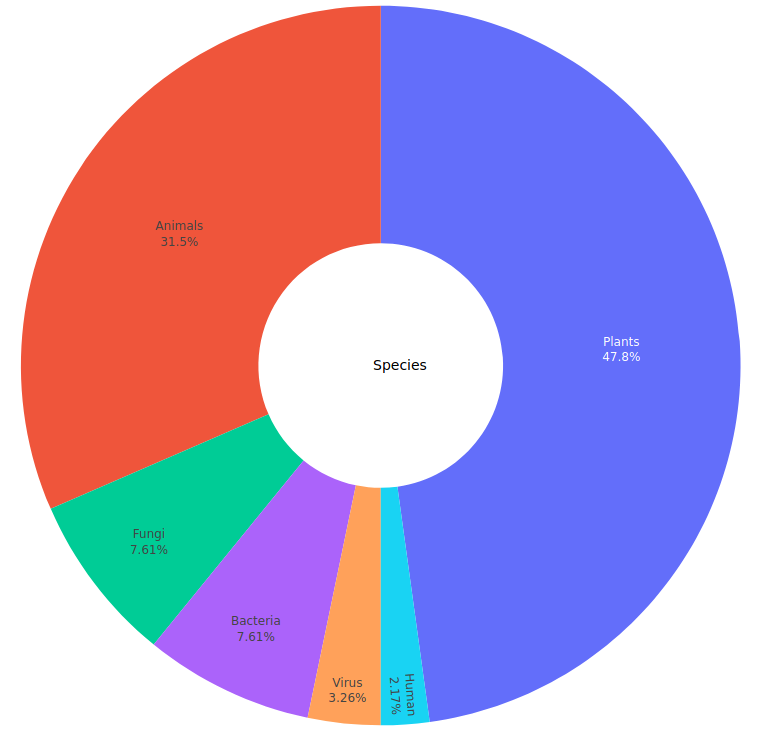
Figure 2: Genomic and transcriptomic projects using LoRDEC (as of 2020-09-01).
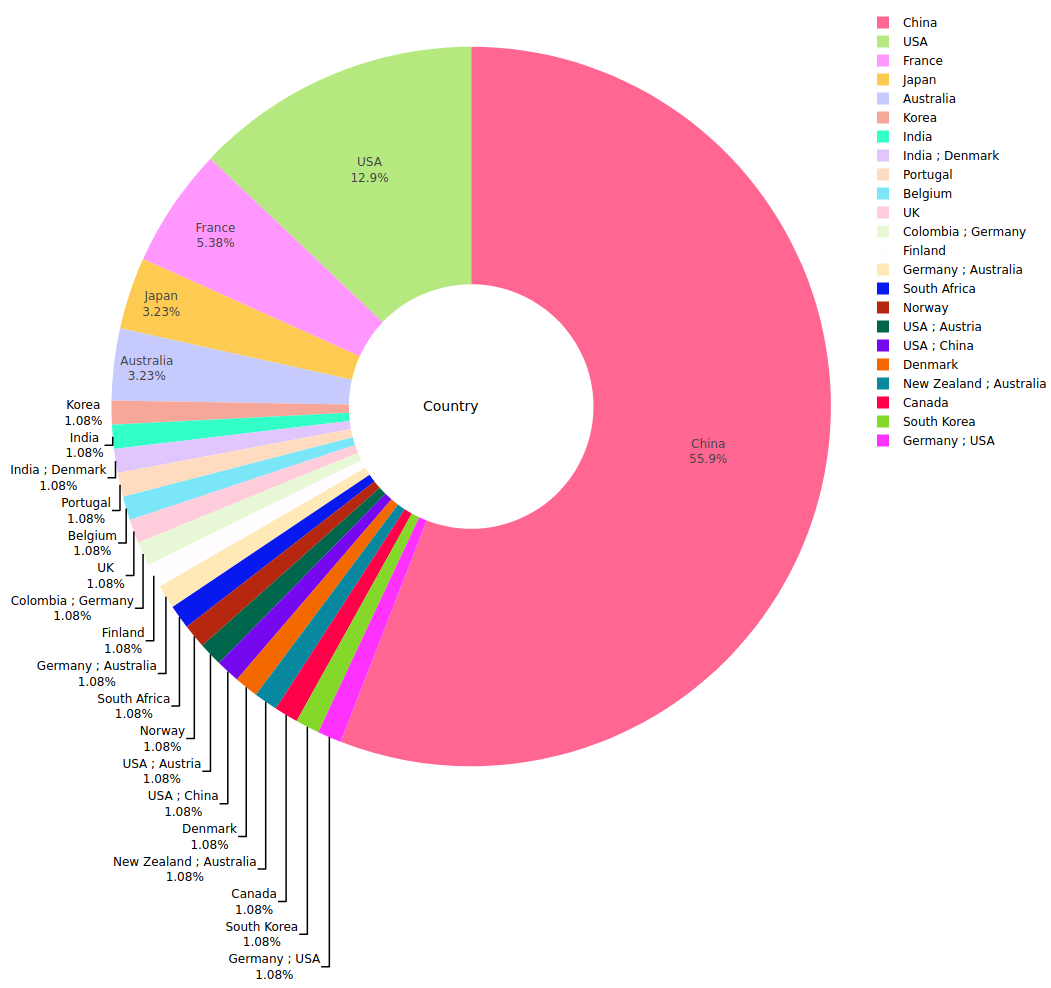
Figure 3: Genomic and transcriptomic projects using LoRDEC (as of 2019-09-01).
Elixir catalog: https://bio.tools/lordec
Frequently Asked Questions (FAQ): it's here.
Publication
The algorithm, the software, and its performances are described in
LoRDEC: accurate and efficient long read error correction
L. Salmela and E. Rivals
Bioinformatics 30(24):3506-3514, 2014, doi: 10.1093/bioinformatics/btu538.
For citations: on G. Scholar, on CrossRef, or on Scopus
Getting the program for Linux or MacOSX
- Official releases page at our gitlab server: https://gite.lirmm.fr/lordec/lordec-releases/wikis/home
- Official page on the ATGC - NGS web platform: http://www.atgc-montpellier.fr/lordec/
- Older versions
- For a stand alone version on the GATB website: http://gatb.inria.fr/software/lordec/
- For interactive use on a Galaxy platform: http://colibread.genouest.org/
- For a version that you can install in your Galaxy platform: https://github.com/genouest/tools-colibread
- For a stand alone version on the GATB website: http://gatb.inria.fr/software/lordec/
Completed genome / transcriptome projects that used LoRDEC
We record completed projects that explicitely used LoRDEC for read correction and are associated with an available publication (as of 01-09-2020)
A total of 93 projects have been completed and published. If you are aware of more send us an email (lordec@lirmm.fr).
Legends for group species: A: animals, B: bacteria, F: fungi, H: human, V: viruses, E: other eukaryotes i.e. protists.
| Number | Species | Type | Journal | DOI | Year | Country | Comments | Speciesgroup |
| 1 | tree Azadirachta indica | genome | G3: Genes, Genomes, Genetics | https://doi.org/10.1534/g3.116.030056 | 2016 | India | evergreen tree, Meliaceae family, medicinal, pesticidal & insecticidal properties; company | P |
| 2 | sorghum Sorghum bicolor BTx623 genotype | transcriptome | Nature Communications | http://dx.doi.org/10.1038/ncomms11706 | 2016 | USA | important bioenergy crop; TAPIS pipeline | P |
| 3 | rabbit Oryctolagus cuniculus | transcriptome | Scientific Reports | https://doi.org/10.1038/s41598-017-08138-z | 2017 | China | provides meat, fur and wool products worldwide | A |
| 4 | strawberry Fragaria vesca | transcriptome | The Plant Journal | https://doi.org/10.1111/tpj.13462 | 2016 / 2017 | China | wild strawberry (diploid) Fragaria vesca, fruit crop of nutrition & economic value | P |
| 5 | fungus Bipolaris cookei | genome | Scientific Reports | https://doi.org/10.1038/s41598-017-17476-x | 2017 | USA | pathogene sorghum | F |
| 6 | butterfly Heliconius erato | genome | Nature Ecology & Evolution | https://doi.org/10.1038/s41559-016-0052 | 2017 | Finland | evolution | A |
| 7 | fungus Stenocarpella maydis | genome | Fungal Biology | https://doi.org/10.1016/j.funbio.2017.08.006 | 2017 | USA | plant pathogenic fungus; causes Diplodia ear rot | F |
| 8 | wheat Triticum aestivum | genome | Scientific Reports | https://doi.org/10.1038/srep44609 | 2017 | China | hexaploid bread wheat, accounts for over 95% of global wheat production | P |
| 9 | cacao Theobroma cacao L. Criollo | genome | BMC Genomics | https://doi.org/10.1186/s12864-017-4120-9 | 2017 | France | with ATGC PF ; economically important fruit tree crop for tropical countries | P |
| 10 | sugarcane Saccharum officinarum S. spontaneum | transcriptome | BMC Genomics | https://doi.org/10.1186/s12864-017-3757-8 | 2017 | Australia | economic importance of sugarcane in sugar and bioenergy production | P |
| 11 | strawberry Fragaria vesca | transcriptome | DNA Research | https://doi.org/10.1093/dnares/dsx038 | 2018 | China | crop | P |
| 12 | fungus Pochonia chlamydosporia | genome secretome | Scientific Reports | https://doi.org/10.1038/s41598-018-19169-5 | 2018 | China | P. chlamydosporia infects eggs and females of economically important plant-parasitic nematodes | F |
| 13 | chicken Gallus gallus domesticus | genome transcriptome | GigaScience | https://doi.org/10.1093/gigascience/giy086 | 2018 | Korea | black native Korean chicken breed Yeonsan Ogye | A |
| 14 | Western poplar Populus trichocarpa | transcriptome | Frontiers in Plant Science | https://doi.org/10.3389/fpls.2018.00005 | 2018 | USA | tree | P |
| 15 | peanut Arachis monticola | genome | GigaScience | https://doi.org/10.1093/gigascience/giy066 | 2018 | China | wild plant; allotetraploid genome | P |
| 16 | herb Sophora japonica | genome | PlOS One | https://doi.org/10.1371/journal.pone.0202485 | 2018 | China | mitochondrial; traditional herb | P |
| 17 | fungus Candida albicans | genome | Genome Biology and Evolution | https://doi.org/10.1093/gbe/evy166 | 2018 | Japan | strain TIMM 1768; company | F |
| 18 | bat Rousettus aegyptiacus | genome | Cell | https://doi.org/10.1016/j.cell.2018.03.070 | 2018 | USA | army; medical; Egyptian Roussete | A |
| 19 | Chinese hamster Cricetulus griseus | genome | Biotechnology and Bioengineering | https://doi.org/10.1002/bit.26722 | 2018 | USA ; Austria | biotech species for exogenous protein production | A |
| 20 | bermudagrass Cynodon dactylon (L.) Pers. | transcriptome | Plant Physiology and Biochemistry | https://doi.org/10.1016/j.plaphy.2018.08.039 | 2018 | China | Cynodon dactylon (L.) Pers.; turfgrass; economic value | P |
| 21 | tree poplar Populus bolleana Populus alba | genome | Plant Biotechnology Journal | https://doi.org/10.1111/pbi.12989 | 2018 | China | var. pyramidalis; economic interest short transformation time | P |
| 22 | bacteria Staphylococcus aureus | genome | Microbiology Resource Announcements | https://doi.org/10.1128/MRA.00892-18 | 2018 | Canada | strains CMRSA3 & CMRSA6; Methicillin-resistant | B |
| 23 | caddisfly Stenopsyche tienmushanensis | genome | GigaScience | https://doi.org/10.1093/gigascience/giy143 | 2018 | China | species widely employed as bio-indicators in water quality assessment and monitoring. | A |
| 24 | jellyfish medusa Tripedalia cystophora | transcriptome | BMC Genomics | https://doi.org/10.1186/s12864-019-5514-7 | 2019 | Denmark | nervous system development, evolution, neuropeptides | A |
| 25 | grape Berry (C. sauvignon) | transcriptome | G3: Genes, Genomes, Genetics | https://doi.org/10.1534/g3.118.201008 | 2019 | USA | economically crucial grape cultivar | P |
| 26 | shrimp Litopenaeus vannamei | transcriptome | Fish & Shellfish Immunology | https://doi.org/10.1016/j.fsi.2019.01.023 | 2019 | China | economically important shrimp for aquaculture industry | A |
| 27 | Crustacea Armadillidium vulgare | genome | Molecular Biology and Evolution | https://doi.org/10.1093/molbev/msz010 | 2019 | France | evolution sex chromosome | A |
| 28 | golden algae Hydrurus foetidus | genome | F1000 Research | https://doi.org/10.12688/f1000research.16734.1 | 2019 | Norway | evolution and ecological roles of chrysophyte algae | P |
| 29 | rapeseed Brassica napus | genome | Plant Biotechnology Journal | https://doi.org/10.1111/pbi.13115 | 2019 | China | B napus (AACC, 2n=38) important oilseed crop grown worldwide | P |
| 30 | Paper Mulberry Broussonetia papyrifera | genome | Molecular Plant | https://doi.org/10.1016/j.molp.2019.01.021 | 2019 | China | papermaking, forage plant | P |
| 31 | grape Carménère | genome | G3: Genes, Genomes, Genetics | https://doi.org/10.1534/g3.119.400030 | 2019 | USA | economically crucial grape cultivar | P |
| 32 | birds Zosterops borbonicus, Z. pallidus | genome | bioRxiv | http://dx.doi.org/10.1101/505610 | 2018 | France | evolution sex chromosome | A |
| 33 | locust Locusta migratoria | transcriptome | RNA Biology | https://doi.org/10.1080/15476286.2019.1602437 | 2019 | China | economically important threat to agriculture | A |
| 34 | Opium poppy Papaver somniferum | transcriptome | Scientific Data | https://doi.org/10.1038/s41597-019-0082-x | 2019 | China | medicinal plants, only commercial resource of morphinan-based painkillers | P |
| 35 | Kaposi sarcoma-associated herpesvirus | genome | J. of Virology | https://doi.org/10.1128/JVI.00630-19 | 2019 | USA | medical | V |
| 36 | pepper Capsicum annuum | transcriptome | The Plant Journal | https://doi.org/10.1111/tpj.14351 | 2019 | China | economically important | P |
| 37 | Human breast cancer | transcriptome | Scientific Reports | https://doi.org/10.1038/s41598-019-42184-z | 2019 | China | health | H |
| 38 | Lotus Nelumbo nucifera | transcriptome | DNA Research | https://doi.org/10.1093/dnares/dsz010 | 2019 | China | important aquatic horticultural plant | P |
| 39 | common morning glory Ipomoea purpurea | exome | Biorxiv | https://doi.org/10.1101/647164 | 2019 | USA | common agricultural weed | P |
| 40 | Red maple Acer rubrum L. | transcriptome | BMC Plant Biology | https://doi.org/10.1186/s12870-019-1850-7 | 2019 | China | economically important | P |
| 41 | Indian mithun Bos frontalis | genome | BMC Genomics | https://doi.org/10.1186/s12864-019-5980-y | 2019 | India ; Denmark | agriculture | A |
| 42 | Esophageal squamous cell carcinoma | transcriptome | Frontiers in Genetics | https://doi.org/10.3389/fgene.2019.00915 | 2019 | China | health | H |
| 43 | Leucobacter | metagenomics | BMC Genomics | https://doi.org/10.1186/s12864-019-6206-z | 2019 | Portugal | metabolic collaboration | B |
| 44 | brown plan hopper Nilaparvata lugens | transcriptome | BMC Genomics | https://doi.org/10.1186/s12864-019-6049-7 | 2019 | China | pest of rice | A |
| 45 | Mycobacterium bovis | genome | BMC Genomics | https://doi.org/10.1186/s12864-019-5909-5 | 2019 | Belgium | tuberculosis vaccine | B |
| 46 | mouse Mus musculus | transcriptome | BMC Genomics | https://doi.org/10.1186/s12864-019-5903-y | 2019 | USA ; China | retinal development | A |
| 47 | giant grouper (Epinephelus lanceolatus | genome | Marine Drugs | https://doi.org/10.3390/md17090503 | 2019 | China | economically important | A |
| 48 | Weissella hellenica | genome | Frontiers in Microbiology | https://doi.org/10.3389/fmicb.2019.01619 | 2019 | Japan | fermenting bacteria | B |
| 49 | yellowthorn Xanthoceras sorbifolium | transcriptome | GigaScience | https://doi.org/10.1093/gigascience/giz070 | 2019 | China | economically important | P |
| 50 | Schistosoma haematobium | genome | GigaScience | https://doi.org/10.1093/gigascience/giz108 | 2019 | Australia | health, human parasite | A |
| 51 | Chinese rhesus macaque Macaca mulatta | transcriptome | Nature Communications | https://doi.org/10.1038/s41467-019-12174-w | 2019 | China | most widely studied nonhuman primate | A |
| 52 | Armadillidium vulgare | genome | Molecular Biology and Evolution | https://doi.org/10.1093/molbev/msz010 | 2019 | France | model species for sex determination and symboiosis in animals | A |
| 53 | mud crab Scylla paramamosain | transcriptome | Aquaculture | https://doi.org/10.1016/j.aquaculture.2019.734447 | 2019 | China | economically important | A |
| 54 | Teratosphaeria gauchensis | genome | IMA Fungus | https://doi.org/10.1186/s43008-019-0013-7 | 2019 | South Africa | plant pathogen | F |
| 55 | seagrass Zostera japonica | transcriptome | Scientific Reports | https://doi.org/10.1038/s41598-019-51176-y | 2019 | China | ecology | P |
| 56 | redlip mullet Liza haematocheila | genome | Frontiers in Genetics | https://doi.org/10.3389/fgene.2019.01246 | 2019 | South Korea | cultivated fish | A |
| 57 | duck Anas platyrhynchos | transcriptome | Scientific Data | https://doi.org/10.1038/s41597-019-0293-1 | 2019 | China | economically important | A |
| 58 | superb fairy-wren Malurus cyaneus | genome | Molecular Ecology Resources | https://doi.org/10.1111/1755-0998.13124 | 2020 | Germany ; Australia | evolution, endemic Australian bird | A |
| 59 | Boehmeria tricuspis | transcriptome | International Journal of Genomics | https://doi.org/10.1155/2019/4025747 | 2019 | China | asexual reproduction | P |
| 60 | Pseudotaxus chienii | transcriptome | International Journal of Molecular Sciences | https://doi.org/10.3390/ijms21124305 | 2020 | China | ecological and economical value | P |
| 61 | pitaya Hylocereus | transcriptome | International Journal of Molecular Sciences | https://doi.org/10.3390/ijms21093288 | 2020 | China | agriculture | P |
| 62 | poplar Populus alba | transcriptome | Frontiers in Genetics | https://doi.org/10.3389/fgene.2020.00048 | 2020 | China | economically important | P |
| 63 | pacific white shrimp Litopenaeus vannamei | transcriptome | Frontiers in Genetics | https://doi.org/10.3389/fgene.2020.00071 and https://doi.org/10.1016/j.fsi.2019.01.023 | 2019 / 2020 | China | economically important | A |
| 64 | Phakopsora pachyrhizi | transcriptome | Physiological and Molecular Plant Pathology | https://doi.org/10.1016/j.pmpp.2020.101464 | 2020 | USA | soybean pathogen | F |
| 65 | crabapple Malus spp. | transcriptome | Plant Physiology and Biochemistry | https://doi.org/10.1016/j.plaphy.2020.01.033 | 2020 | China | gardening and landscaping | P |
| 66 | Cupressus chengiana | transcriptome | Molecular Ecology | https://doi.org/10.1111/mec.15407 | 2020 | China | ecology | P |
| 67 | human adenovirus type 5 | transcriptome | Communications Biology | https://doi.org/10.1038/s42003-020-0849-9 | 2020 | UK | health | V |
| 68 | oilseed rape Brassica napus | transcriptome | The Plant Journal | https://doi.org/10.1111/tpj.14754 | 2020 | China | economically important | P |
| 69 | alpine grass Stipa capillacea and Stipa purpurea | transcriptome | Journal of Experimental Botany | https://doi.org/10.1093/jxb/eraa194 | 2020 | China | evolution | P |
| 70 | Rhododendron delavayi | genome | PeerJ | https://doi.org/10.7717/peerj.9048 | 2020 | China | gardening and landscaping | P |
| 71 | ramie Boehmeria nivea L. Gaudich | transcriptome | Industrial Crops and Products | https://doi.org/10.1016/j.indcrop.2020.112544 | 2020 | China | economically important | P |
| 72 | red palm weevil Rhynchophorus ferrugineus | transcriptome | PeerJ | https://doi.org/10.7717/peerj.9133 | 2020 | China | palm tree pest | A |
| 73 | thraustochytrid Aurantiochytrium limacinum | genome | Data in Brief | https://doi.org/10.1016/j.dib.2020.105729 | 2020 | France | industrial production | E |
| 74 | Penium margaritaceum | transcriptome | Cell | https://doi.org/10.1016/j.cell.2020.04.019 | 2020 | USA | evolution | P |
| 75 | razor clam Sinonovacula constricta | transcriptome | Frontiers in Genetics | https://doi.org/10.3389/fgene.2020.00664 | 2020 | China | economically important | A |
| 76 | Litchi Litchi chinensis Sonn. | transcriptome | Postharves Biology and Technology | https://doi.org/10.1016/j.postharvbio.2020.111252 | 2020 | China | economically important | P |
| 77 | hybrid: blunt snout bread x topmouth culter Megalobramaamblycephala x Culter alburnus | transcriptome | BMC Genomics | https://doi.org/10.1186/s12864-020-06866-7 | 2020 | China | maternal effects on transcription | A |
| 78 | Armillaria gallica 012m | genome | Brazilian Journal of Microbiology | https://doi.org/10.1007/s42770-020-00317-x | 2020 | China | plant disease | B |
| 79 | kuruma shrimp Marsupenaeus japonicus | transcriptome | Fish and Shellfish Immunology | https://doi.org/10.1016/j.fsi.2020.07.018 | 2020 | China | economically important | A |
| 80 | tall fescue Festuca arundinacea | transcriptome | BMC Plant Biology | https://doi.org/10.1186/s12870-020-02572-4 | 2020 | China | agriculture | P |
| 81 | ticks Acari: Ixodidae | genome | Cell | https://doi.org/10.1016/j.cell.2020.07.023 | 2020 | China | health | A |
| 82 | soil bacteria Mesorhizobium | genome | Microbial Genomics | https://doi.org/10.1099/mgen.0.000426 | 2020 | New Zealand ; Australia | nitrogen binding | B |
| 83 | human breast cancer | transcriptome | bioRxiv | https://doi.org/10.1101/2020.07.15.199851 | 2020 | Japan | health | H |
| 84 | macadamia Macadamia integrifolia | genome | bioRxiv | https://doi.org/10.1101/2020.05.25.114009 | 2020 | Australia | agriculture | P |
| 85 | Tripterygium wilfordii | transcriptome | bioRxiv | https://doi.org/10.1101/2020.06.29.176958 | 2020 | China | traditional chinese medicine, medical value | P |
| 86 | Ascosphaera apis | transcriptome | bioRxiv | https://doi.org/10.1101/2020.03.19.998625 | 2020 | China | fungal disease of honeybees | F |
| 87 | C. elegans microbiome | metagenomics | bioRxiv | https://doi.org/10.1101/2020.04.22.055426 | 2020 | Germany ; USA | model species microbiome | B |
| 88 | Hard Fescue Turfgrass Festuca brevipila | transcriptome | bioRxiv | https://doi.org/10.1101/2020.02.26.966952doi | 2020 | USA | gardening and landscaping | P |
| 89 | Stevia rebaudiana | transcriptome | Research Square | https://doi.org/10.21203/rs.3.rs-24957/v2 | 2020 | China | economically important | P |
| 90 | Sea vomit Didemnum vexillum | genome | Research Square | https://doi.org/10.21203/rs.3.rs-23360/v1 | 2020 | Colombia ; Germany | evolution | A |
| 91 | Liriodendron chinense (Hemsl.) Sarg. | transcriptome | Research Square | https://doi.org/10.21203/rs.3.rs-32884/v1 | 2020 | China | economically important | P |
| 92 | Gnetum | transcriptome | Research Square | https://doi.org/10.21203/rs.3.rs-36058/v1 | 2020 | China | economically important | P |
| 93 | wild apple Malus sieversii | transcriptome | Research Square | https://doi.org/10.21203/rs.3.rs-47953/v1 | 2020 | China | plant disease | P |
Contacts
- To contact us about LoRDEC please use: lordec@lirmm.fr
- We have a list to contact regular users: lordec-users@lirmm.fr
Authors
- Leena Salmela Department of Computer Science, P. O. Box 68, FIN-00014 University of Helsinki, Finland
- Eric Rivals LIRMM and Institut de Biologie Computationelle, CNRS and Université Montpellier, Montpellier, France
Contributors: Julien Veyssier LIRMM, CNRS and Université Montpellier, Montpellier, France
Acknowledgements and supports
We thank :
- Erwan Drezen (GATB team)
- Yvan Le Bras (GenOuest platform)
- Vincent Lefort (LIRMM ATGC plaftorm)
- Cyril Monjeaud (Plate-form GenOuest)
- Dominique Lavenier (IRISA Rennes)
- all authors of GATB library
- Amal Makrini (LIRMM ATGC plaftorm)
Supports:
- Institut de Biologie Computationelle http://www.ibc-montpellier.fr/
- LIRMM http://www.lirmm.fr
- Institut Français de Bioinformatique through funding of 3GENSEQ
- France Génomique
- ANR Colib'read (ANR-12-BS02-0008) http://colibread.inria.fr/
- Academy of Finland [grant number 267591]
- Défi MASTODONS SePhHaDe: http://www.lirmm.fr/mastodons/
- CNRS http://www.cnrs.fr/
- Université Montpellier http://www.umontpellier.fr/
- University of Helsinki http://www.cs.helsinki.fi/
 |
 |
 |
 |