Overview
Metagenomics is a sequencing based method for probing biodiversity in uncultured environments, like sea water, coalmine soil, gut, or water supply pipes. Each experiment yields a huge number of sequencing reads, and a read is part of the DNA signature of some species. With phylogenetic placement, i.e., the bioinformatic task aiming at guessing from which species a read come from, one can interrogate the biodiversity in a sample of interest.
The presence of viruses or bacteria can be monitored using phylogenetic placement. We aim at designing very efficient tools for performing the phylogenetic placement of sequencing reads accurately. We target especially viruses in our applications.
Remark This project was launched before the Covid-19 outbreak.
Results and outcomes
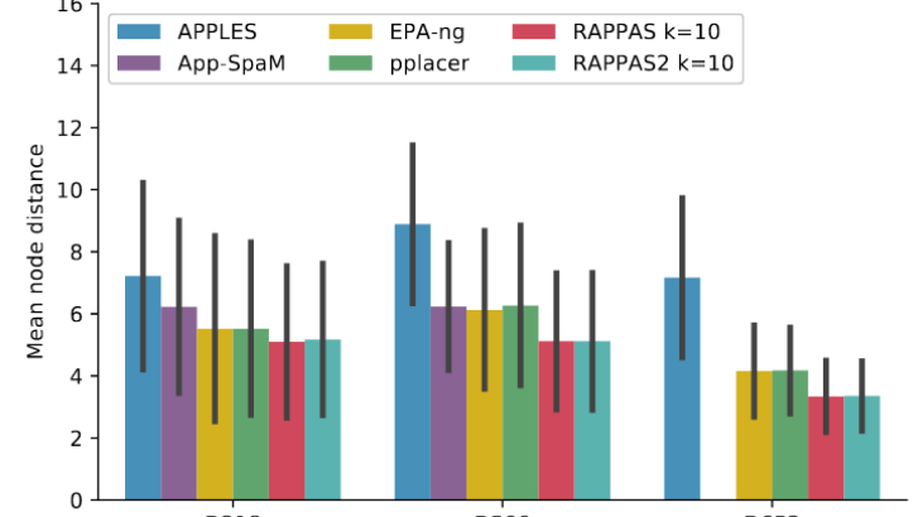
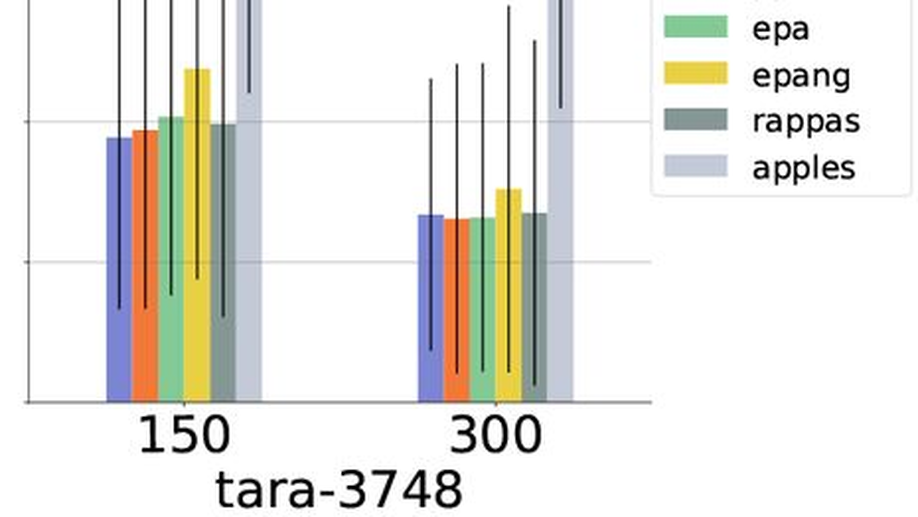
- PEWO is a benchmark framework to test and evaluate novel and existing phylogenetic placement methods. It can be easily extended to incorporate new methods and to run tests automatically.
- SHERPAS is a tool to detect recombination between known virus strains of a given species.
- RAPPAS is a fast and alignment free software for phylogenetic placement.
Team
- Fabio Pardi
- Benjamin Linard
- Nikolay Romashchenko
- Guillaume Scholz