This year, several French researchers asked me to participate to the jury of their “Habilitation” defence. The habilitation is the diploma that officially entitled somebody to supervise PhD students. Here, are a few notes about these defences. Nota Bene: all illustrations come from their manuscripts, published papers, and/or websites.
First, Dr. Hugues Richard defended his habilitation in Applied Mathematics and Statistics on Feb. 14, 2022 in Paris Sorbonne University. He presented a review of his work on statistical methods for analysing high throughput sequencing data, and transcriptomics data. As a reviewer, I appreciated the variety of techniques he employed to devise computational solutions that are widely applicable in practise. Besides that, the entire jury discovered his epistemological views on the utility, or better to say the relevance of “Big data” for genomics.
Title: Statistical methods for analysing high throughput sequencing data (pdf)
Author: Hugues RICHARD, Sorbonne University, Paris, and Robert Koch Institute, Berlin.
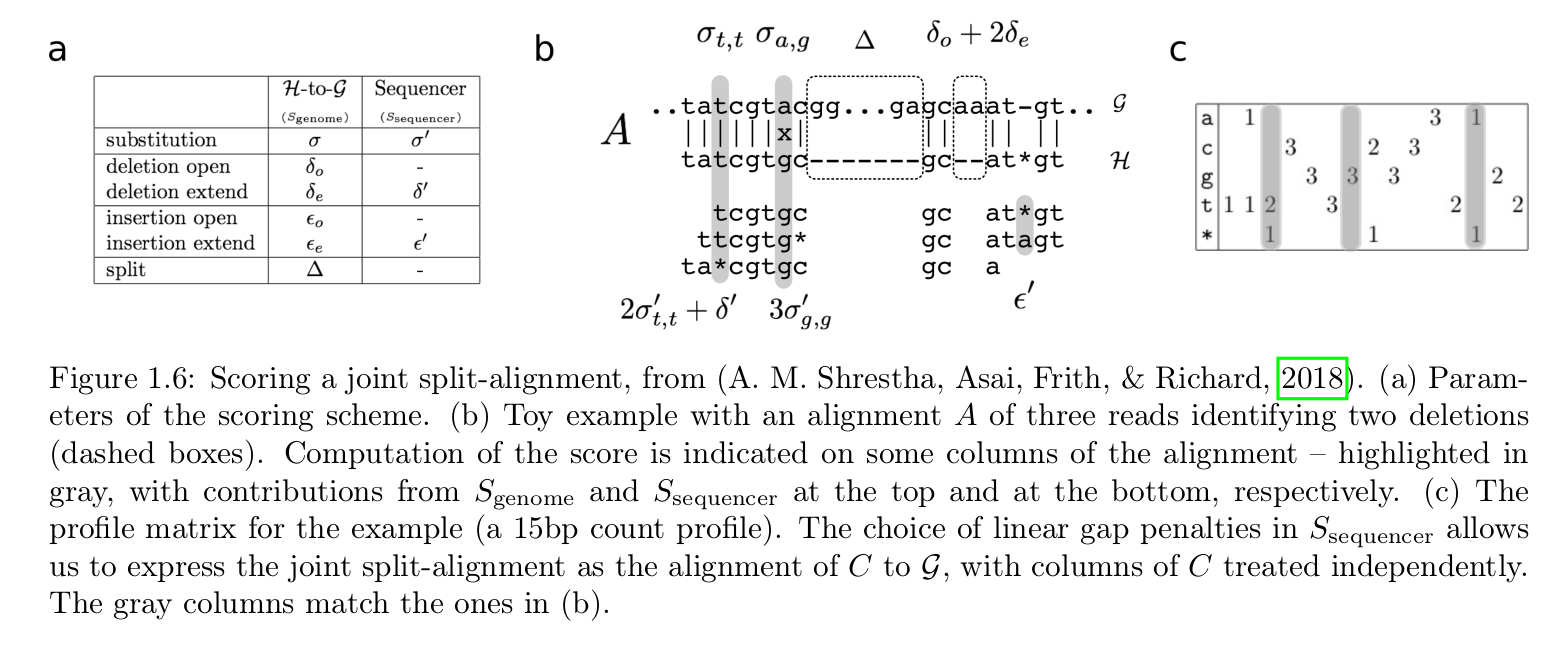
Fig 1.6 from H. Richard habilitation manuscript: it illustrates a method to score a split-alignment of a read.
Recently, on April 4, 2022 in Montpellier, I served as the “president” of the jury during the habilitation in Evolutionary Biology of Dr. Anna-Sophie Fiston-Lavier, who presented an overview of her investigations about the impact of transposable elements on genome evolution. Transposable elements, are pieces of the genome that were brought to light by Barbara McClintock in the 60s (who later earned a Nobel price in 1983 for her discovery – one of the few Nobel price female winners in Physiology/Medicine). Anna-Sophie made significant contributions on methods to discover and annotate transposable elements in sequenced genomes, especially in plants and insects. This task remains dauting given that, for some plant genomes, up to 90% of it is composed of transposable elements. The jury also emphasised Anna-Sophie’s endeavour and efforts to promote science among girls/women at both school and university levels.
Title: Transposable elements: impact on the structure and evolution of genomes
Author: Anna-Sophie Fiston-Lavier, Montpellier University.
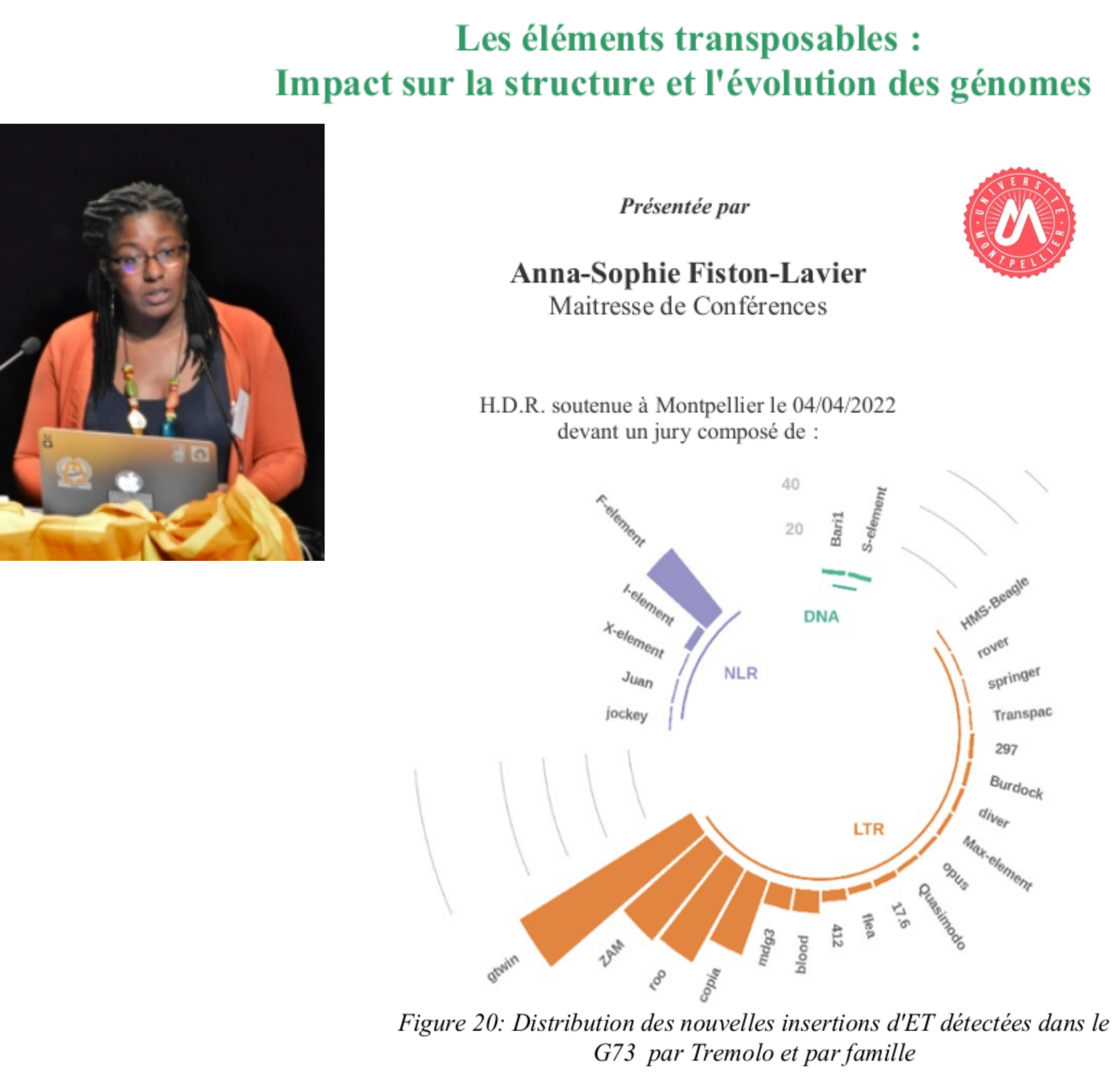
Composed image including the front matter of Dr Fiston manuscript, a photo of her, a figure from this manuscript about the distribution of TE insertions into subfamilies
In Toulouse University, on April 13, 2022, Dr. Matthias Zytnicki, defended his habilitation in Computer Science about computational methods to analyse the sequencing of small RNAs. Also more recent than that of transposable elements, the discovery of small RNAs is acknowledged as a major breakthrough in molecular biology and also was celebrated by the attribution of a Nobel Price in 2006. Matthias report about computational challenges of analysing sequencing data of small RNAs shows very well that important differences, compared to classical transcriptomic data, pose specific problems. Dr Zytnicki has tackled these issues and proposed original algorithms for solving them. This clearly suggests that analyses that were done with methods and parameters intended for messenger RNA data sets, could be reanalysed in more relevant way with methods adapted to small RNAs, and provide new results, new insights on the role of the latter.
Title: Computational methods for analysing sequencing data on small RNAs
Author: Matthias Zytnicki, INRAE Toulouse.
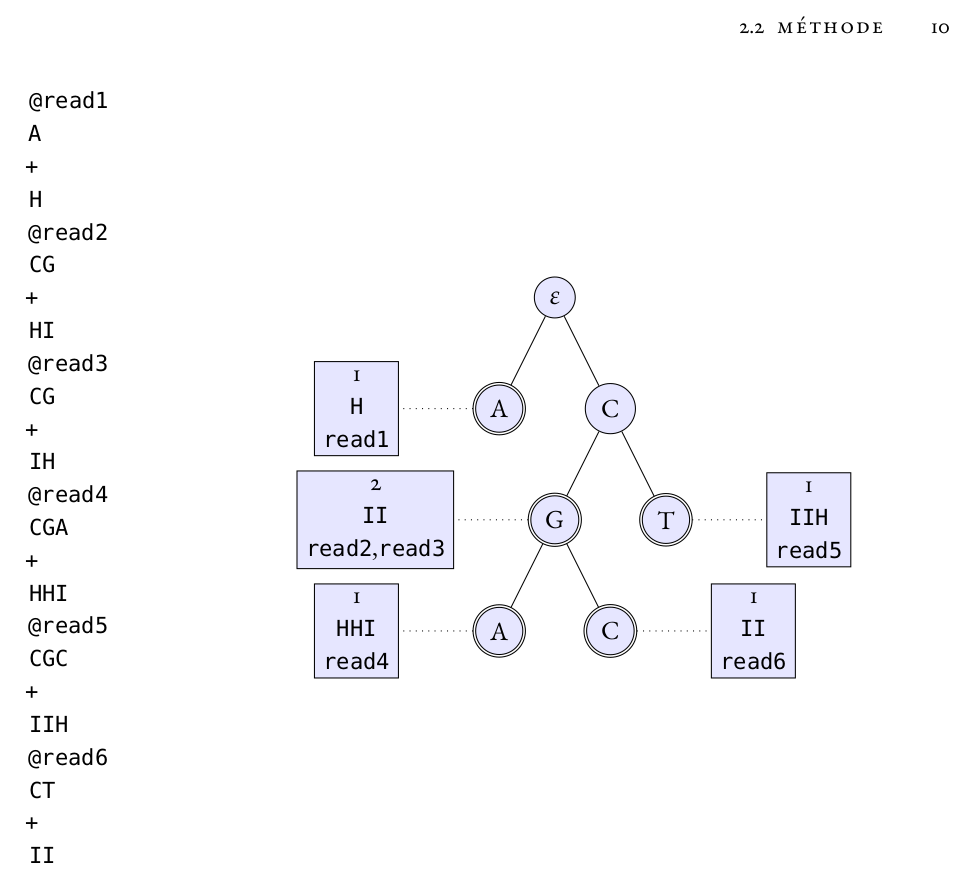
Figure 2.1 on page 10 of Dr Zytnicki’s habilitation manuscript. It illustrates a read mapping algorithm that is well suited for small RNAs
Again, I truly want congratulate all three of them, and to thank them again for asking me to belong to their jury: reading their manuscript and listening to their presentation taught me a lot, and certainly was an opportunity to keep up to date with state of the art in their respective domain of computational biology.
Take a look at their work!
