A long term collaboration with a Physics Lab of Montpellier, more specifically, with the Biophysics team, and a molecular biology team from the Institute of Functional Genomics, delivers a first publication on a new model for the kinetics of ribosomes on messenger RNAs.
We are glad to announce that the preprint, which is already available on ArXiV, is now accepted in PlOS Computational Biology. For full reference of this publication visit the publication page publication page
To get an overview and a summary, please take a look at the poster that was presented at the Annual Meeting of the International Physics of Living Systems (iPoLS) Network, in May 2022.
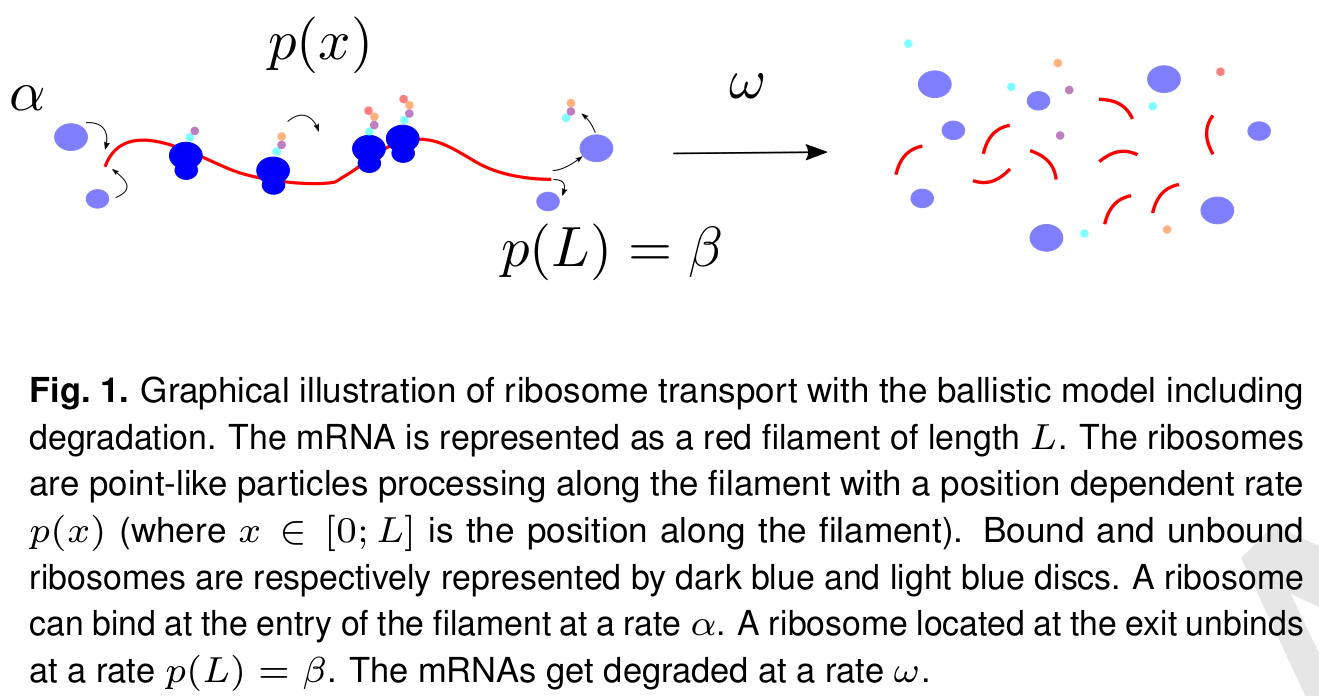
In cells, ribosomes perform the translation of messenger RNA (mRNA) molecules into proteins. However, several ribosomal complexes can read and process simultaneously the same linear RNA molecule, but cannot overtake each other. The position of ribosomes on all mRNA can be “registered” by Ribosome sequencing (a.k.a. Ribo-seq) followed by intensive bioinformatic analyses. Our new ballistic model incorporates the fact distinct RNA are processed by different number of ribosomes and also the degradation of RNA molecules (i.e., the fact that each mRNA has a limited lifetime). It takes advantage of these to infer kinetic parameters.
Evaluation of the model performance require to exploit the Human polysome sequecing data produced at IGF and analyzed at by my team at the LIRMM.
Congrats and thanks to all collaborators.